|
|
||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||
 |
Prof YMD Lo |
Modern developments in molecular biology have provided many powerful tools for molecular analysis and research. While much of the past research has relied on the use of nucleic acids extracted from cells, research is now moving towards plasma to explore the biological and diagnostic implications of nucleic acids in this cell-free portion of the blood.
The first finding of circulating DNA was published in 1947, when Mandel and Métais used chemical methods to demonstrate the presence of DNA and RNA in the plasma of both healthy and sick individuals. This finding is particularly impressive since it occurred before Watson and Crick discovered the structure of the double helix. However, it was not until the 1960s that interest in this area re-emerged, when circulating DNA was found in patients with systemic lupus erythematosus. Further development took place in the 1970s when concentrations of circulating DNA were found to be increased in patients with cancer.1 In particular, levels of circulating DNA are higher in patients with metastatic disease and decrease after successful treatment. Due to the technological limitations at the time, researchers were unable to elucidate the genetic origin of circulating DNA in cancer patients, and were therefore unable to tell whether the DNA may have been released by the tumour cells or from other tissues in the body.
In 1989, Stroun et al proposed that some of the DNA found in the circulation of patients with cancer may be released from the tumour cells, based on the fact that this DNA had certain biochemical characteristics of neoplastic cells.2 The conclusive proof of this hypothesis came 5 years later when tumour-derived oncogene mutations were detected in the plasma and serum of patients with cancer.3,4 The spectrum of mutations closely matched those of the primary tumour. This research provides good evidence to suggest that, through an as yet undefined process, tumour cells release DNA into the circulation, which then circulates as plasma tumour DNA (Figure 1).
The turning point for this research came in 1996, when Chen et al5 and Nawroz et al5 reported the presence of microsatellite shift in the plasma and serum of patients with cancer, which correlated with that in the primary tumour. Similarly, a loss of an allele in the primary tumour correlated with the loss of the same allele in the plasma and serum. This work has caught the attention of researchers, and the field has rapidly expanded during the past few years.
Figure 1. Release of tumour DNA into the circulation.
References

1. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-650.
2. Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318-322.
3. Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71.
4. Vasioukhin V, Anker P, Maurice P, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodys-plastic syndrome or acute myelogenous leukaemia. Br J Haematol 1994;86:774-779.
5. Chen XQ, Stroun M, Magnenat JL, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med 1996;2:1033-1035.
6. Nawroz H, Koch W, Anker P, et al. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med 1996;2:1035-1037.
Tumour-derived Genetic Alterations in Plasma and Serum
|
|
Dr P Anker University of Geneva Geneva, Switzerland |
Circulating DNA

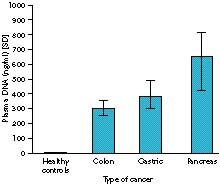
In 1977, radioimmunoassay showed that there was more circulating DNA in patients with cancer than in healthy people.1 In addition, some cancers were associated with higher levels of DNA than others, with pancreatic cancer being associated with the highest levels. It was later found that early lung cancer was associated with less circulating DNA than advanced lung cancer, but more than healthy people.2
Meanwhile, Fournie et al found that the survival rate was longer for patients with small amounts of circulating DNA than for patients with more than 100 ng/ml of circulating DNA.3 Maebo determined a cut off point of 80 ng/ml to detect 71% of patients with cancer and 37% with benign tumours (Table 1).2 This research also showed that plasma DNA levels tend to increase following unsuccessful treatment, and decrease following successful treatment.1,2
Table 1. Cancer detection rate according to plasma DNA level of 80 ng/ml.
| Condition | Number of patients | Detection rate (%) |
| Healthy control | 59 | 0 |
| Benign tumour | 54 | 37 |
| Cancer | 45 | 71 |
Ras Mutations

Since Sorenson identified K-ras mutations associated with colon and pancreatic tumours,4 several researchers have investigated Ras mutations in plasma DNA in myeloid disorders and colon and pancreatic cancers.
There are 2 methods for identifying Ras mutations in circulating DNA. The first is PASA PCR (allele-specific assay polymerase chain reaction), in which mutation specific primers are used, and only genes corresponding to the primer are amplified. The second method is RFLP PCR, in which a restriction enzyme cleavage site is created by making a 1-base substitution on codon 11. Wild type genes are digested, but mutated genes are not, and can therefore be separated by electrophoresis. Interestingly, a mutation in serum DNA was found in 4 of 7 patients diagnosed with pancreatitis. These 4 patients were diagnosed with a malignant pancreatic tumour 5 to 14 months later, indicating that the presence of a tumour can be detected before it can be clinically diagnosed.
p53 Mutations

Research into plasma p53 mutations has not been as successful as that for Ras mutations, possibly because the technique for detecting p53 is less sensitive. However, Gonzalez et al found that 37% of patients with small cell lung cancer had p53 mutations in plasma.5 In another study, 8 of 12 patients with breast cancer and 7 of 12 patients with lymphoma were harbouring p53 mutations in the plasma. Dr Anker suggested that it is therefore worth pursuing this approach.
Microsatellite Alterations

Many researchers have investigated microsatellite DNA alterations. Microsatellite DNA is unstable in cancer cells and the alterations appear as new alleles or allele expansions of LOH. Microsatellite DNA alterations are a part of neoplastic progression and may serve as clinical markers.
Summary of Ras Mutations in Plasma or Serum
|
Figure 1. Plasma DNA levels in gastrointestinal malignancy quantitated by radioimmunoassay.
Plasma, tumour, and lymphocyte DNA are used as PCR templates and compared. One primer is labelled radioactively or with a fluorescent marker and PCR products are separated by denaturing gel electrophoresis. After autoradiography or laser measurement, the alleles of tumour and plasma DNA are compared with the alleles of the lymphocyte DNA, which is used as a control.
To summarise the research into microsatellite alterations to date, some markers appear to be useful as prognostic markers, although sometimes no clinical correlation has been found. Markers can be used for follow-up, since there is a correlation with tumour response and disappearance of abnormal plasma DNA and with tumour progression and reappearance of abnormal plasma DNA. Some markers allow for a high rate of tumour detection. Surprisingly, clean LOH can be found in the plasma serum, although the results are variable.
Circulating RNA

Circulating RNA has been found in the plasma or serum. This is important since frequent mRNA overexpression can be found in cancer cells and several genes have been found in the plasma of patients with cancer studies performed to date involve melanoma, and pancreatic, breast, colorectal, and lung cancers. RNA is absent in the serum of most healthy donors. However, circulating RNA is partly a quantitative factor and real time PCR may improve the detection rate.
Conclusions

Plasma or serum DNA of patients with cancer show characteristics similar to those of the primary tumour. Tumour-related RNA has also been found in the serum of patients with cancer. Plasma DNA may serve as a prognostic factor for certain cancers, as can microsatellite alterations and Ras mutations, and may be used for monitoring the effects of treatment.
References

1. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-650.
2. Maebo A. Plasma DNA level as a tumor marker in primary lung cancer [article in Japanese]. Nihon Kyobu Shikkan Gakkai Zasshi 1990;28:1085-1091.
3. Fournie GJ, Courtin JP, Laval F, et al. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett 1995;91:221-227.
4. Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71.
5. Gonzalez R, Silva JM, Sanchez A, et al. Microsatellite alterations and TP53 mutations in plasma DNA of small-cell lung cancer patients: follow-up study and prognostic significance. Ann Oncol 2000;11:1097-1104.
Nuclear Genetic Changes Drive Progression of Human Cancers
|
|
Prof D Sidransky Johns Hopkins University School of Medicine Baltimore, USA |
Among the most promising markers for cancer diagnosis are microsatellite alterations, methylation, mitochondria, and viral DNA. Microsatellites are highly polymorphic markers that show that DNA in serum or plasma share multiple genetic changes with the primary tumour and therefore cannot be derived from any source other than the primary cancer. However, the sensitivity of microsatellite alterations in serum is not high a serious limitation for a general diagnostic tool. Tissue-specific methylation is a new type of methylation and is a promising area for research.
Mitochondrial DNA

Nuclear mutations of mitochondrial DNA are thought to drive the tumour process. Prof Sidransky described the research into the functional significance of mitochondria and how they arise in the cell and become homoplasmic. All the cellular mitochondria appear to share the same mutations. There are between 10 to 100 mitochondria in a normal cell, with estimates of 10- to 100-fold higher in cancer cells, which forms the basis for research into diagnostic approaches.
Specific mitochondrial mutations can be identified in up to 50% of patients with certain types of cancer, including head and neck, bladder, and lung cancer.1 Many of the mutations are in the D-loop region, which is involved in replication, but it is unclear whether they are functionally significant. However, some tumours have more than one mutation. The enrichment of mitochondria in tumour cells and saliva in head and neck cancer and the increased copy number of mitochondria make them attractive as tumour markers.
In lung cancer, 40% of tumours have primary mutations, many of them in the D-loop. However, few of the mutations are coding mutations, and there are multiple mutations in the same tumour. Again, the mitochondria are homoplasmic.
The ligation-specific assay identified specific mutations in the mitochondria following bronchoalveolar lavage. Amplification of the target DNA from p53 or mitochondrial DNA enabled quantification of mitochondrial versus nuclear mutations to ascertain the amount of enrichment. This research showed very dilute nuclear mutation, while approximately 30% of mitochondrial clones contained mutation, suggesting 100-fold enrichment justifying its potential use as a diagnostic tool. Almost all of the mitochondrial mutations were non-coding mutations, although 2 to 3% are coding mutations and almost all of these are found in respiratory complex I. These functional mutations can give rise to oxygen radicals, which may lead to mutated phenotypes in tumour cells.
The question arose as to how the mutations became homoplasmic. A specific sequence was identified in the D310 area in mitochondrial DNA of lung cancer. The frequency of mitochondrial frameshift mutations rapidly rose when different tumour types were investigated with a simple PCR assay (Table 1). Overall, 24% of tumours investigated had a specific mutation in the D310 region, enabling further research into how and when the mutations arise. There are now several examples of immediate homoplasmic expansion of mitochondrial mutations in severe dysplasia or carcinoma in situ, suggesting that they occur early in the tumour progression. Once mitochondrial mutations occur, they rapidly become homoplasmic.
In order to examine the normal distribution of frameshift mutations in constitutive DNA, amplicons of the regions from lymphocytes of patients who had tumours that expressed specific deletions or insertions were cloned. Fifteen of 61 patients had a frameshift alteration, suggesting that these mitochondrial mutations are due to selection of pre-existing mutations.
Table 1. Mitochondrial mutations in different tumour types.
| Tumour type | Frameshift mutation (%) |
| Gastric | 62 |
| Head and neck | 41 |
| Breast | 29 |
| Colorectal |
28 |
| Lung | 22 |
| Bladder | 20 |
| Ovarian | 0 |
| Prostate | 0 |
Viral DNA in Serum

Human papilloma virus (HPV) is one of the most promising viral markers in serum or plasma. A study of 70 patients with head and neck cancer who were investigated for paired tumour and sera samples showed that 13 patients had HPV-16-positive tumours. Eleven of the 13 patients (85%) had HPV DNA in the serum, and all 11 patients had stage III or IV disease, suggesting that there is some propensity for identifying this marker in patients with advanced disease. Six of the 11 patients (55%) had detectable levels of circulating cell-free HPV DNA.
The research into HPV DNA was extended to cervical cancer. 292 patients were included in an analysis, some of whom had already been treated for cervical cancer 175 had invasive cervical cancer, 57 had carcinoma in situ, and 60 were healthy controls. All patients were examined for HPV 16 or 18 E7 DNA by conventional and real time quantitative PCR assays.
Only 14 of 292 patients were positive for HPV 16 or 18 E7 DNA, of whom 12 had invasive cervical cancer, 1 had carcinoma in situ and 1 was in the control group. There was a good correlation for HPV in the plasma and the primary tumour, with 9 women having HPV in the plasma. Six of the 9 women had HPV 16 in the genital tract, 1 had evidence of HPV 18, and 1 had evidence of another type of HPV. Only 1 woman with HPV DNA in the plasma had no evidence of HPV in the genital tract. Despite the good correlation of presence of HPV in the plasma and the primary tumour, it would be interesting to know why there was a low rate of circulating DNA abnormalities among these patients.
Promoter Methylation Changes

Promoter methylation can turn off genes and can be a valuable marker for tumour suppressor gene inactivation. A recent study analysed APC methylation in tissue samples from 99 patients with lung cancer. Ninety five patients had methylation changes in the tissues, and there was some correlation with survival. Forty two of 89 patients (47%) had positive plasma or serum samples, indicating a good correlation between plasma or serum and tissue samples. However, there was no significance between APC hypermethylation level in the serum or plasma and any clinical data. Interestingly, this marker is seen at high levels in tumour tissues, and is high in the serum or plasma but is never seen in control patients, making this a potential universal marker.
References

1. Fliss MS, Usadel H, Caballero OL, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 2000;287:2017-2019.
Home | Current Issue | Back Issues | Congress Calendar
Free Subscription | Editorial Board |